Research
We’re interested in a lot of things, and we’ve always got some side projects going on. Some of those turn into main projects eventually. We do a lot of methods development, meaning a mix of theory, software, and data analysis. We run a dry lab, meaning we use mostly whiteboards and keyboards instead of test tubes and pipettes, and try not to spill tea on our computers. Here’s some of our main threads of research:
Spatial Population Genetics
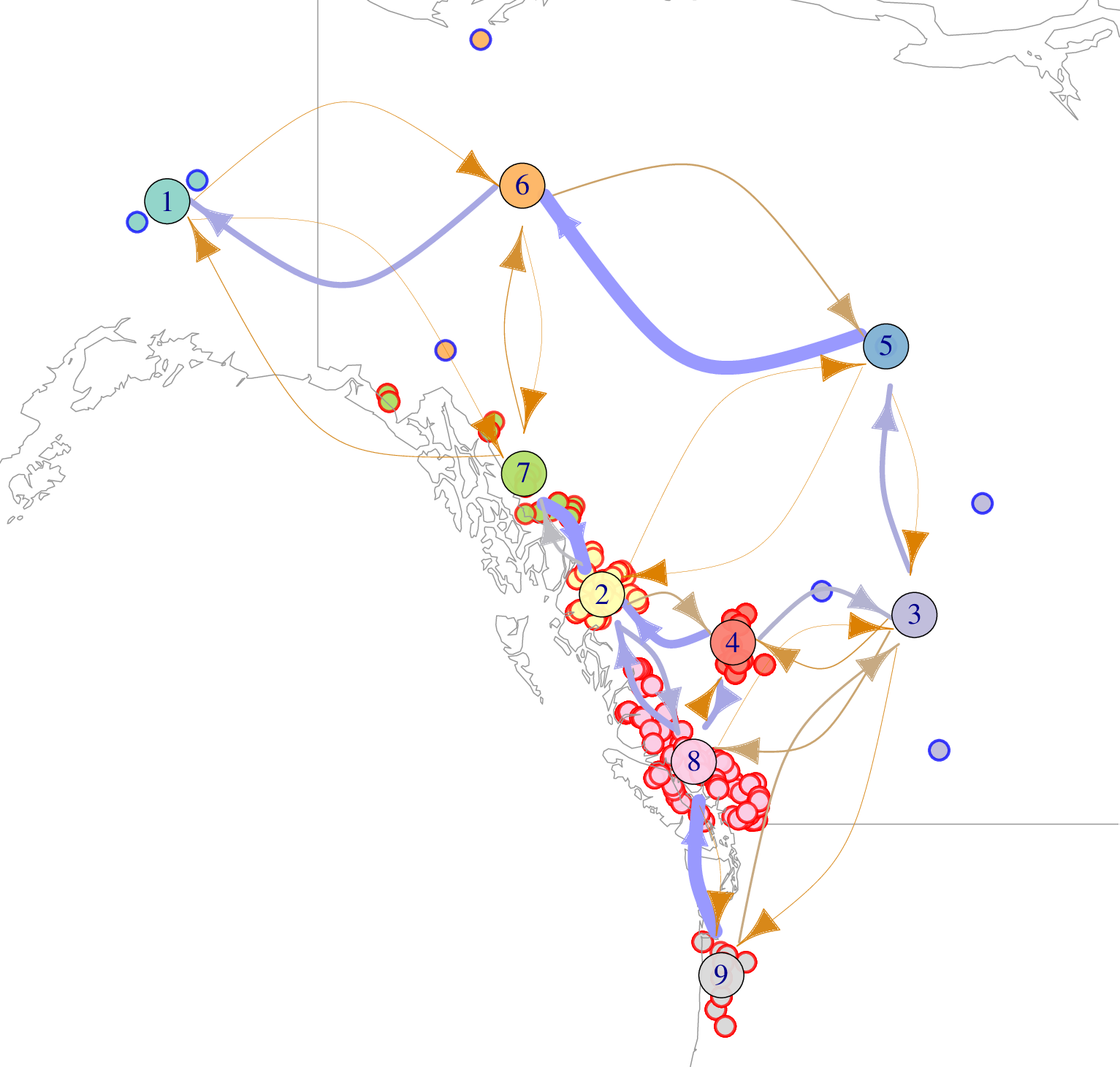
The world is big, and organisms don’t all live in the same place (happily). Geography can be a problem for popgen methods that make simpler assumptions, but - more excitingly - is a promising source of information when tied with modern georeferenced genomic data. Where’d that genome come from, on a map? How far away from each other do close relatives tend to live? What’s a map of population density look like? How do organisms (or, pollen, or seeds) move around on the map? We’re working on these and other questions, using various tools from math to machine learning, as well as on good ways to simulate populations that live, reproduce, die, and evolve across interesting and realistic geographies.
Machine learning approaches to population genetics
Computers are getting faster nearly as fast as our ability to sequence lots of genomes, and this has made it possible to write software tools that act more and more like our brains: as abstract methods to learn how to do seemingly arbitrary tasks. Just like computers can tell funny pictures of dogs from boring pictures of cats, we’re using machine learning methods to learn things about genomes, like distinguishing bits that are under recent natural selection from bits that aren’t or to locate genomes on a map. Doing this well takes a lot of know-how, of course: understanding what information to put in, how to make it available, and what questions to ask determines the result as much here as it does in the machine learning methods cropping up in the rest of life.
Understanding the influence of selection on genomic variation
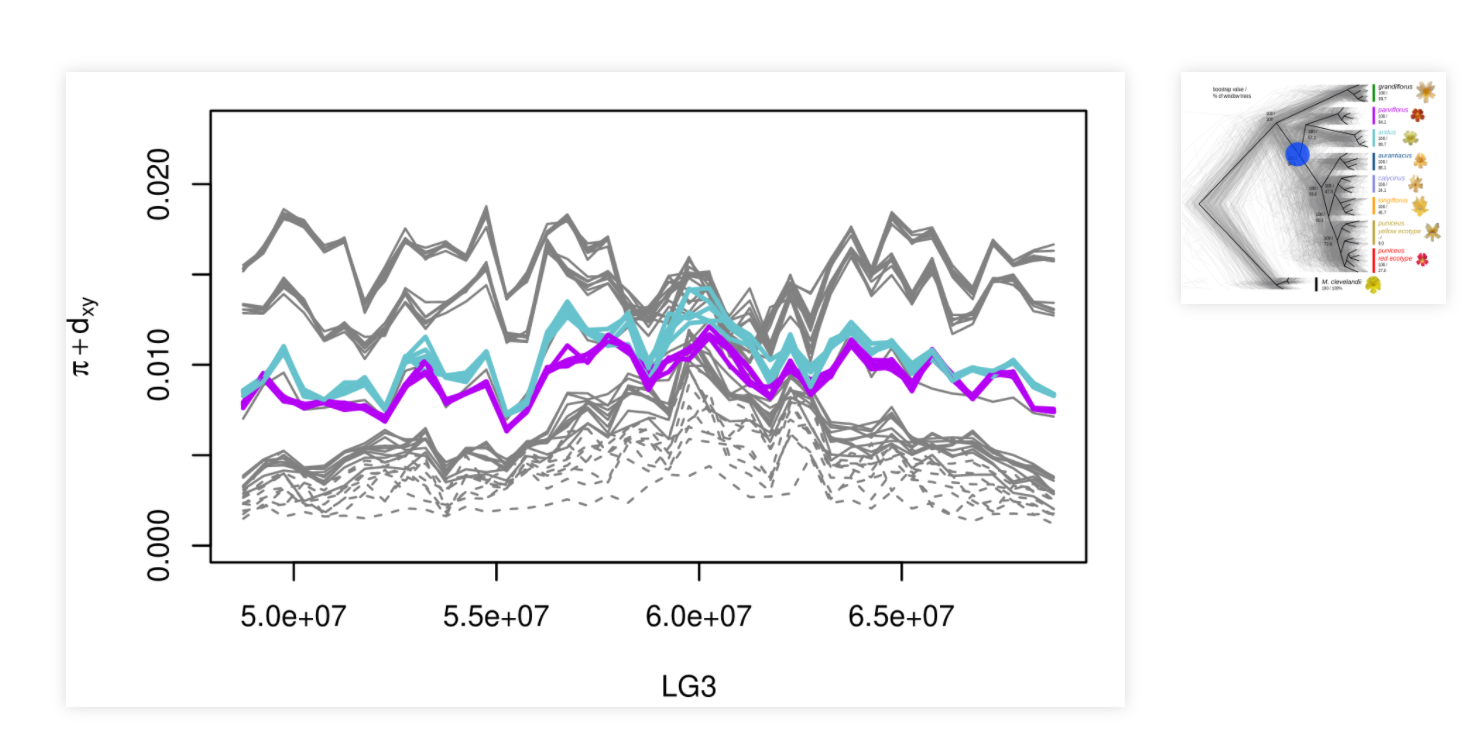
We understand really quite well how evolution works in a lot of ways, especially on a microscopic level: we know a lot about how natural selection acts on heritable traits and the underlying genetic variation. But there’s a lot of outstanding questions of scale: How much does natural selection act? How strongly does it constrain organisms’ genomes? Is natural selection mostly keeping organisms near a fitness peak, or is it constantly moving different traits around, maybe even in different directions in different places? A unifying question here is: How much is genetic variation influenced by selection as opposed to drift (i.e., random genealogical noise). We’re working on this by developing methods to identify the action of selection, theory to describe the process of adaptation, and by plain old fashioned digging into the data.
Simulation methods
Fundamental to lots of our work are simulations: in studying complex things like how natural selection affects big genomes, or how big populations evolve across geography, it’s important to be able to try out different situations and see what happens. And, simulations are the basic ingredient to lots of modern machine learning methods (i.e., simulation-based inference). It’s challenging to make a good simulation that reasonably realistically describes the various parts of reality we’d like to study, from organisms moving around and interacting down to the details of how DNA is inherited. So, we put a lot of time in collaborative work developing these tools that we and many other people use:
- We’re core members ofthe PopSim consortium,
- as well as the tskit group,
- and contribute regularly to SLiM.